- ImageCLEF 2025
- LifeCLEF 2025
- ImageCLEF 2024
- LifeCLEF 2024
- ImageCLEF 2023
- LifeCLEF 2023
- ImageCLEF 2022
- LifeCLEF2022
- ImageCLEF 2021
- LifeCLEF 2021
- ImageCLEF 2020
- LifeCLEF 2020
- ImageCLEF 2019
- LifeCLEF 2019
- ImageCLEF 2018
- LifeCLEF 2018
- ImageCLEF 2017
- LifeCLEF2017
- ImageCLEF 2016
- LifeCLEF 2016
- ImageCLEF 2015
- LifeCLEF 2015
- ImageCLEF 2014
- LifeCLEF 2014
- ImageCLEF 2013
- ImageCLEF 2012
- ImageCLEF 2011
- ImageCLEF 2010
- ImageCLEF 2009
- ImageCLEF 2008
- ImageCLEF 2007
- ImageCLEF 2006
- ImageCLEF 2005
- ImageCLEF 2004
- ImageCLEF 2003
- Publications
- Old resources
You are here
ImageCLEFmedical Caption
Welcome to the 6th edition of the Caption Task!
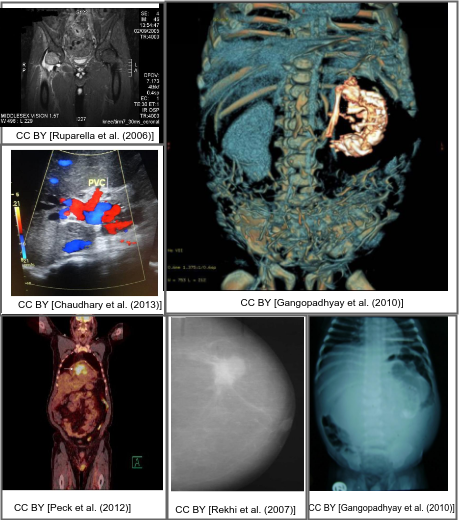
Motivation
Interpreting and summarizing the insights gained from medical images such as radiology output is a time-consuming task that involves highly trained experts and often represents a bottleneck in clinical diagnosis pipelines.
Consequently, there is a considerable need for automatic methods that can approximate this mapping from visual information to condensed textual descriptions. The more image characteristics are known, the more structured are the radiology scans and hence, the more efficient are the radiologists regarding interpretation. We work on the basis of a large-scale collection of figures from open access biomedical journal articles (PubMed Central), as well as radiology images from original medical cases. All images in the training data are accompanied by UMLS concepts extracted from the original image caption.
Lessons learned:
- In the first and second editions of this task, held at ImageCLEF 2017 and ImageCLEF 2018, participants noted a broad variety of content and situation among training images. In 2019, the training data was reduced solely to radiology images, with ImageCLEF 2020 adding additional imaging modality information, for pre-processing purposes and multi-modal approaches
- The focus in ImageCLEF 2021 lay in using real radiology images annotated by medical doctors. This step aims at increasing the medical context relevance of the UMLS concepts
- For ImageCLEF 2022, an extended version of the ImageCLEF 2020 dataset is used
- To reduce the scope and size of concepts, several concept extraction tools are analyzed prior to caption pre-processing methods.
- Concepts with less occurrence will be removed
- As uncertainty regarding additional source was noted, we will clearly separate systems using exclusively the official training data from those that incorporate additional sources of evidence
News
- 15.11.2021: website goes live
- 11.01.2022: AICrowd challenges online
- 17.01.2022: Training and validation datasets released
- 15.03.2022: Test dataset released
- 06.05.2022: Run submission deadline
- 13.05.2022: Release of the processed results
Task Description
In ImageCLEFmedical Caption 2022 consists of two substaks:
- Concept Detection Task: https://www.aicrowd.com/challenges/imageclef-2022-caption-concept-detection
- Caption Prediction Task: https://www.aicrowd.com/challenges/imageclef-2022-caption-caption-predic...
Concept Detection Task
The first step to automatic image captioning and scene understanding is identifying the presence and location of relevant concepts in a large corpus of medical images. Based on the visual image content, this subtask provides the building blocks for the scene understanding step by identifying the individual components from which captions are composed. The concepts can be further applied for context-based image and information retrieval purposes.
Evaluation is conducted in terms of set coverage metrics such as precision, recall, and combinations thereof.
Caption Prediction Task
On the basis of the concept vocabulary detected in the first subtask as well as the visual information of their interaction in the image, participating systems are tasked with composing coherent captions for the entirety of an image. In this step, rather than the mere coverage of visual concepts, detecting the interplay of visible elements is crucial for strong performance.
Evaluation of this second step is based on metrics such as BLEU that have been designed to be robust to variability in style and wording. In this year we will evaluate other potential metrics like METEOR, ROUGE, and CIDEr.
Data
A subset of the extended Radiology Objects in COntext (ROCO) dataset [1], for this edition without imaging modality information, is used for both subtasks. As in previous editions, the dataset originates from biomedical articles of the PMC OpenAccess subset.
Training Set: Consists of 83,275 radiology images
Validation Set: Consists of 7,645 radiology images
Test Set: Consists of 7,601 radiology images
Concept Detection Task
The concepts were generated using a reduced subset of the UMLS 2020 AB release, which includes the sections (restriction levels) 0, 1, 2, and 9. To improve the feasibility of recognizing concepts from the images, concepts were filtered based on their semantic type. Concepts with very low frequency were also removed, based on suggestions from previous years.
Caption Prediction Task
For this task each caption is pre-processed in the following way:
- Numbers and words containing numbers were removed.
- All punctuation was removed.
- Lemmatization was applied using spaCy.
- Captions were converted to lower-case.
Evaluation methodology
Concept Detection
Evaluation is conducted in terms of F1 scores between system predicted and ground truth concepts, using the following methodology and parameters:
- The default implementation of the Python scikit-learn (v0.17.1-2) F1 scoring method is used. It is documented here.
- A Python (3.x) script loads the candidate run file, as well as the ground truth (GT) file, and processes each candidate-GT concept sets
- For each candidate-GT concept set, the y_pred and y_true arrays are generated. They are binary arrays indicating for each concept contained in both candidate and GT set if it is present (1) or not (0).
- The F1 score is then calculated. The default 'binary' averaging method is used.
- All F1 scores are summed and averaged over the number of elements in the test set (7'601), giving the final score.
The ground truth for the test set was generated based on the same reduced subset of the UMLS 2020 AB release which was used for the training data (see above for more details).
NOTE : The source code of the evaluation tool is available here. To execute it, first change the variables ground_truth_path and submission_file_path in evaluator.py (making sure that both files do not have header rows!), then either run data/test_runs/docker/docker_run.sh on a Linux system with docker installed, or run python3 evaluator.py after installing the requirements in requirements.txt on Python 3.6+.
Caption Prediction
This year, in addition to the BLEU scores, ROUGE is used as a secondary metric. Other metrics like METEOR and CIDR will be reported after the challenge concludes. For evaluation, each caption will also be pre-processed (similar to the pre-processing steps mentioned above). The BLEU scores are calculated using the following methodology and parameters:
- The default implementation of the Python NLTK (v3.2.2) (Natural Language ToolKit) BLEU scoring method is used. It is documented here and based on the original article describing the BLEU evaluation method
- A Python (3.6) script loads the candidate run file, as well as the ground truth (GT) file, and processes each candidate-GT caption pair
- Each caption is pre-processed in the following way:
- The caption is converted to lower-case
- All punctuation is removed an the caption is tokenized into its individual words
- Stopwords are removed using NLTK's "english" stopword list
- Lemmatization is applied using spacy's Lemmatizer
- The BLEU score is then calculated. Note that the caption is always considered as a single sentence, even if it actually contains several sentences. No smoothing function is used.
- All BLEU scores are summed and averaged over the number of captions, giving the final score.
NOTE : The source code of the evaluation tool is available here (no longer available). To execute it, first change the variables ground_truth_path and submission_file_path in evaluator.py (making sure that both files do not have header rows!), then either run data/test_runs/docker/docker_run.sh on a Linux system with docker installed, or run python3 evaluator.py after installing the requirements in requirements.txt on Python 3.7+.
The ROUGE scores are calculated using the following methodology and parameters:
- The native python implementation of ROUGE scoring method is used. It is designed to replicate results from the original perl package that was introduced in the original article describing the ROUGE evaluation method.
- Specifically, we calculate the ROUGE-1 (F-measure) score, which measures the number of matching unigrams between the model-generated text and a reference.
- A Python (3.7) script loads the candidate run file, as well as the ground truth (GT) file, and processes each candidate-GT caption pair
- Each caption is pre-processed in the following way:
- The caption is converted to lower-case
- Stopwords are removed using NLTK's "english" stopword list
- Lemmatization is applied using spacy's Lemmatizer
- The ROUGE score is then calculated. Note that the caption is always considered as a single sentence, even if it actually contains several sentences.
- All ROUGE scores are summed and averaged over the number of captions, giving the final score.
Participant registration
Please refer to the general ImageCLEF registration instructions
Preliminary Schedule
- 15 November 2021: Registration opens
- 17 January 2022: Release of the training and validation sets
- 14 March 2022: Release of the test sets
- 22 April 2022: Registration closes
- 6 May 2022: Run submission deadline
- 13 May 2022: Release of the processed results by the task organizers
- 27 May 2022: Submission of participant papers [CEUR-WS]
- 27 May – 13 June 2022: Review process of participant papers
- 13 June 2022: Notification of acceptance
- 1 July 2022: Camera ready copy of participant papers and extended lab overviews [CEUR-WS]
- 5-8 September 2022: CLEF 2022, Bologna, Italy
Submission Instructions
The submissions will be received through the AIcrowd system.
Please note that each group is allowed a maximum of 10 runs per subtask.
Concept Detection
For the submission of the concept detection task we expect the following format:
- <Figure-ID>|<Concept-ID-1>;<Concept-ID-2>;<Concept-ID-n>
You need to respect the following constraints:
- The separator between the figure ID and the concepts has to be a pipe character ( | )
- The separator between the UMLS concepts has to be a semicolon (;)
- Each figure ID of the test set must be included in the submitted file exactly once (even if there are not concepts)
- The same concept cannot be specified more than once for a given figure ID
- The maximum number of concepts per image is 100
Caption prediction
For the submission of the caption prediction task we expect the following format:
- <Figure-ID>|<description>
You need to respect the following constraints:
- The separator between the figure ID and the description has to be a pipe charachter ( | )
- Each figure ID of the testset must be included in the runfile exactly once
- You should not include special characters in the description.
Results
The tables below contain only the best runs of each team, for a complete list of all runs please see the following CSV files:
- Concept Detection Task: concept_best_run_secondary.csv, concept_all_runs_secondary.csv
- Caption Prediction Task: caption_best_run.csv, caption_all_runs.csv
Concept Detection Task
For the cconcept detection task, the ranking is based on the F1 score as described in the Evaluation Methodologies section above. Additionally, a Secondary F1 score was calculated using a subset of manually validated concepts (anatomy and image modality) only.
| Group Name | Best Run | F1 Score | Secondary F1 | Rank |
|---|---|---|---|---|
| AUEB-NLP-Group | 182358 | 0.451123 | 0.790724 | 1 |
| fdallaserra | 182324 | 0.450532 | 0.822240 | 2 |
| CSIRO | 182343 | 0.447134 | 0.793645 | 3 |
| eecs-kth | 181750 | 0.436013 | 0.854593 | 4 |
| vcmi | 182097 | 0.432871 | 0.863373 | 5 |
| PoliMi-ImageClef | 182296 | 0.431955 | 0.851217 | 6 |
| SSNSheerinKavitha | 181995 | 0.418433 | 0.654361 | 7 |
| IUST_NLPLAB | 182307 | 0.398086 | 0.673188 | 8 |
| Morgan_CS | 182150 | 0.351988 | 0.628052 | 9 |
| kdelab | 182346 | 0.310428 | 0.411958 | 10 |
| SDVA-UCSD | 181691 | 0.307932 | 0.552432 | 11 |
Caption Prediction Task
For the caption prediction task, the ranking is based on the BLEU scores. Additional metrics were mainly calculated using the pycocoevalcap [pypi.org] library.
- For the METEOR score, the v1.5 jar is used for the calculation, which was introduced in Meteor Universal: Language Specific Translation Evaluation for Any Target Language [aclanthology.org]
- CIDEr score is based on CIDEr: Consensus-Based Image Description Evaluation [cv-foundation.org]
- The java implementation of SPICE introduced in SPICE.pdf [panderson.me] is used.
- For the calculation of the BERTScore, the official implementation Tiiiger/bert_score: BERT score for text generation [github.com] is utilized, which is based on BERTScore: Evaluating Text Generation with
BERT (openreview.net). As a pretrained model to calculate the score, microsoft/deberta-xlarge-mnli was used.
| Group Name | Best Run | BLEU | ROUGE | METEOR | CIDEr | SPICE | BERTScore | Rank |
|---|---|---|---|---|---|---|---|---|
| IUST_NLPLAB | 182275 | 0.482796 | 0.142206 | 0.092754 | 0.030356 | 0.007231 | 0.561192 | 1 |
| AUEB-NLP-Group | 181853 | 0.322165 | 0.166498 | 0.073744 | 0.190217 | 0.031258 | 0.598860 | 2 |
| CSIRO | 182268 | 0.311356 | 0.197439 | 0.084137 | 0.269338 | 0.046228 | 0.623419 | 3 |
| vcmi | 182325 | 0.305778 | 0.173754 | 0.074579 | 0.204668 | 0.035778 | 0.604429 | 4 |
| eecs-kth | 182337 | 0.291664 | 0.115669 | 0.062403 | 0.131695 | 0.021826 | 0.572823 | 5 |
| fdallaserra | 182342 | 0.291316 | 0.201163 | 0.081942 | 0.256383 | 0.046359 | 0.610078 | 6 |
| kdelab | 182351 | 0.278265 | 0.158389 | 0.073531 | 0.411406 | 0.051164 | 0.600314 | 7 |
| Morgan_CS | 182238 | 0.254931 | 0.144074 | 0.055924 | 0.148146 | 0.023232 | 0.583494 | 8 |
| MAI_ImageSem | 182105 | 0.221136 | 0.184723 | 0.067541 | 0.251316 | 0.039311 | 0.605873 | 9 |
| SSNSheerinKavitha | 182248 | 0.159522 | 0.042500 | 0.022645 | 0.016949 | 0.007213 | 0.545069 | 10 |
CEUR Working Notes
For detailed instructions, please refer to this PDF file. A summary of the most important points:
- All participating teams with at least one graded submission, regardless of the score, should submit a CEUR working notes paper.
- Teams who participated in both tasks should generally submit only one report
- Submission of reports is done through EasyChair – please make absolutely sure that the author (names and order), title, and affiliation information you provide in EasyChair match the submitted PDF exactly!
- Strict deadline for Working Notes Papers: 27 May 2022 (23:59 CEST)
- Strict deadline for CEUR-WS Camera Ready Working Notes Papers: 1 July 2022 (23:59 CEST)
- Make sure to include the signed Copyright Form
- Templates are available here
- Working Notes Papers should cite both the ImageCLEF 2022 overview paper as well as the ImageCLEFmedical task overview paper, citation information is available in the Citations section below.
CLEF 2022
- CLEF 2022 Website
- Programme
- Poster stands: Poster sessions can allocate up to 20 posters. The dimensions of the available stands 200cm x 100cm, vertical. Notice that hanging posters directly from the wall is NOT allowed
ImageCLEFmedical at CLEF 2022
-
S1 - Tuesday 6 September
17:20-18:50 (GMT+2)
ImageCLEF Overview of Tasks (5 x 15 minutes)- [1] “Overview of ImageCLEFmedical 2022 – Caption Prediction and Concept Detection”,
Raphael Brüngel, University of Applied Sciences and Arts Dortmund, Germany (physical)
- [1] “Overview of ImageCLEFmedical 2022 – Caption Prediction and Concept Detection”,
-
S2 - Wednesday 7 September
08:50-10:20 (GMT+2)
ImageCLEF Medical: Caption & Tuberculosis (7 x 14 minutes)- [1] “CSIRO at ImageCLEFmedical Caption 2022”,
Bowen Xin, Australian e-Health Research Centre, Commonwealth Scientific and Industrial Research Organisation, Australia (physical) - [2] “Detecting Concepts and Generating Captions from Medical Images: Contributions of the VCMI Team to ImageCLEFmedical 2022 Caption”,
Isabel Rio-Torto, University of Porto, Portugal (physical) - [3] “CMRE-UoG team at ImageCLEFmed Caption 2022 Task: Concept Detection and Image Captioning”,
Francesco Dalla Serra, University of Glasgow, Glasgow, UK (physical) - [4] “IUST_NLPLAB at ImageCLEFmedical Caption Tasks 2022”,
Mohammad Mahdi Javid, Iran University of Science and Technology, Islamic Republic of Iran (physical) - [5] “CSIRO at the ImageCLEFmed 2022 Tuberculosis Caverns Detection Challenge: A 2D and 3D Deep Learning Detection Network Approach”,
Hang Min, Australian e-Health Research Centre, Commonwealth Scientific and Industrial Research Organisation, Australia (physical)
- [1] “CSIRO at ImageCLEFmedical Caption 2022”,
-
Posters - Wednesday 7 September
13:30-15:00 (GMT+2)- [1] “Detecting Concepts and Generating Captions from Medical Images: Contributions of the VCMI Team to ImageCLEFmedical 2022 Caption”,
Isabel Rio-Torto, University of Porto, Portugal - [2] "NeuralDynamicsLab at ImageCLEF Medical 2022",
Georgios Moschovis, KTH Royal Institute of Technology, Stockholm
- [1] “Detecting Concepts and Generating Captions from Medical Images: Contributions of the VCMI Team to ImageCLEFmedical 2022 Caption”,
-
S3 - Wednesday 7 September
17:20-18:50 (GMT+2)
ImageCLEF Coral & Fusion (4 x 15 minutes)- [4] “ImageCLEF 2022 final remarks and discussions“
For more details, see ImageCLEF 2022
Citations
When referring to ImageCLEF 2022, please cite the following publication:
- Bogdan Ionescu, Henning Müller, Renaud Péteri, Johannes Rückert,
Asma Ben Abacha, Alba G. Seco de Herrera, Christoph M. Friedrich,
Louise Bloch, Raphael Brüngel, Ahmad Idrissi-Yaghir, Henning Schäfer,
Serge Kozlovski, Yashin Dicente Cid, Vassili Kovalev, Liviu-Daniel
Ștefan, Mihai Gabriel Constantin, Mihai Dogariu, Adrian Popescu,
Jérôme Deshayes-Chossart, Hugo Schindler, Jon Chamberlain, Antonio
Campello, Adrian Clark, Overview of the ImageCLEF 2022: Multimedia
Retrieval in Medical, Social Media and Nature Applications, in
Experimental IR Meets Multilinguality, Multimodality, and Interaction.
Proceedings of the 13th International Conference of the CLEF
Association (CLEF 2022), Springer Lecture Notes in Computer Science
LNCS, Bologna, Italy, September 5-8, 2022. -
@inproceedings{ImageCLEF2022,
author = {Bogdan Ionescu and Henning M\"uller and Renaud P\’{e}teri
and Johannes R\"uckert and Asma {Ben Abacha} and Alba G. Seco
de Herrera and Christoph M. Friedrich and Louise Bloch and Raphael
Br\"ungel and Ahmad Idrissi-Yaghir and Henning Sch\"afer and Serge
Kozlovski and Yashin Dicente Cid and Vassili Kovalev and Liviu-Daniel
\c{S}tefan and Mihai Gabriel Constantin and Mihai Dogariu and Adrian
Popescu and J\'er\^ome Deshayes-Chossart and Hugo Schindler and Jon
Chamberlain and Antonio Campello and Adrian Clark},
title = {{Overview of the ImageCLEF 2022}: {Multimedia Retrieval in
Medical, Social Media and Nature Applications}},
booktitle = {Experimental IR Meets Multilinguality, Multimodality, and
Interaction},
series = {Proceedings of the 13th International Conference of the CLEF
Association (CLEF 2022)},
year = 2022,
volume = {},
publisher = {{LNCS} Lecture Notes in Computer Science, Springer},
pages = {},
month = {September 5-8},
address = {Bologna, Italy}
}
When referring to ImageCLEFmedical 2022 Caption general goals, general results, etc. please cite the following publication which will be published by September 2022:
- Johannes Rückert, Asma Ben Abacha, Alba G. Seco de Herrera, Louise Bloch, Raphael Brüngel, Ahmad Idrissi-Yaghir, Henning Schäfer, Henning Müller and Christoph M. Friedrich. Overview of ImageCLEFmedical 2022 – Caption Prediction and Concept Detection, in Experimental IR Meets Multilinguality, Multimodality, and Interaction. CEUR Workshop Proceedings (CEUR-WS.org), Bologna, Italy, September 5-8, 2022.
- BibTex:
@inproceedings{ImageCLEFmedicalCaptionOverview2022,
author = {R\"uckert, Johannes and Ben Abacha, Asma and G. Seco de Herrera, Alba and Bloch, Louise and Br\"ungel, Raphael and Idrissi-Yaghir, Ahmad and Sch\"afer, Henning and M\"uller, Henning and Friedrich, Christoph M.},
title = {Overview of {ImageCLEFmedical} 2022 -- {Caption Prediction and Concept Detection}},
booktitle = {CLEF2022 Working Notes},
series = {{CEUR} Workshop Proceedings},
year = {2022},
volume = {},
publisher = {CEUR-WS.org },
pages = {},
month = {September 5-8},
address = {Bologna, Italy}
}
Contact
- Join our mailing list: https://groups.google.com/d/forum/imageclefcaption
- Follow our ResearchGate project: https://www.researchgate.net/project/ImageCLEF-2022-ImageCLEFmedical-Caption
- Follow @imageclef
- Johannes Rückert <johannes.rueckert(at)fh-dortmund.de>, University of Applied Sciences and Arts Dortmund, Germany
- Asma Ben Abacha <abenabacha(at)microsoft.com>, Microsoft, USA
- Alba García Seco de Herrera <alba.garcia(at)essex.ac.uk>,University of Essex, UK
- Christoph M. Friedrich <christoph.friedrich(at)fh-dortmund.de>, University of Applied Sciences and Arts Dortmund, Germany
- Henning Müller <henning.mueller(at)hevs.ch>, University of Applied Sciences Western Switzerland, Sierre, Switzerland
- Louise Bloch <louise.bloch(at)fh-dortmund.de>, University of Applied Sciences and Arts Dortmund, Germany
- Raphael Brüngel <raphael.bruengel(at)fh-dortmund.de>, University of Applied Sciences and Arts Dortmund, Germany
- Ahmad Idrissi-Yaghir <ahmad.idrissi-yaghir(a)fh-dortmund.de>, University of Applied Sciences and Arts Dortmund, Germany
- Henning Schäfer <henning.schaefer(at)uk-essen.de>, University of Applied Sciences and Arts Dortmund, Germany
Organizers:
Acknowledgments
[1] O. Pelka, S. Koitka, J. Rückert, F. Nensa und C. M. Friedrich „Radiology Objects in COntext (ROCO): A Multimodal Image Dataset“, Proceedings of the MICCAI Workshop on Large-scale Annotation of Biomedical data and Expert Label Synthesis (MICCAI LABELS 2018), Granada, Spain, September 16, 2018, Lecture Notes in Computer Science (LNCS) Volume 11043, Page 180-189, DOI: 10.1007/978-3-030-01364-6_20, Springer Verlag, 2018.

