- ImageCLEF 2025
- LifeCLEF 2025
- ImageCLEF 2024
- LifeCLEF 2024
- ImageCLEF 2023
- LifeCLEF 2023
- ImageCLEF 2022
- LifeCLEF2022
- ImageCLEF 2021
- LifeCLEF 2021
- ImageCLEF 2020
- LifeCLEF 2020
- ImageCLEF 2019
- LifeCLEF 2019
- ImageCLEF 2018
- LifeCLEF 2018
- ImageCLEF 2017
- LifeCLEF2017
- ImageCLEF 2016
- LifeCLEF 2016
- ImageCLEF 2015
- LifeCLEF 2015
- ImageCLEF 2014
- LifeCLEF 2014
- ImageCLEF 2013
- ImageCLEF 2012
- ImageCLEF 2011
- ImageCLEF 2010
- ImageCLEF 2009
- ImageCLEF 2008
- ImageCLEF 2007
- ImageCLEF 2006
- ImageCLEF 2005
- ImageCLEF 2004
- ImageCLEF 2003
- Publications
- Old resources
You are here
ImageCLEFmed Tuberculosis
Motivation
Welcome to the 6th edition of the Tuberculosis Task!
Tuberculosis (TB) is a bacterial infection caused by a germ called Mycobacterium tuberculosis. About 130 years after its discovery, the disease remains a persistent threat and a leading cause of death worldwide according to WHO. This bacteria usually attacks the lungs, but it can also damage other parts of the body. Generally, TB can be cured with antibiotics. However, the different types of TB require different treatments, and therefore the detection of the TB type and characteristics are important real-world tasks.
Task history and lessons learned:
- Tuberculosis task exists in ImageCLEF since 2017 and it was modified from year to year.
- In the first and second editions of this task, held at ImageCLEF 2017 and ImageCLEF 2018, participants had to detect Multi-drug resistant patients (MDR subtask) and to classify the TB type (TBT subtask) both based only on the CT image. After 2 editions we concluded that the MDR subtask was not possible to solve based only on the image. In the TBT subtask, there was a slight improvement in 2018 with respect to 2017 on the classification results, however, not enough considering the amount of extra data provided in the 2018 edition, both in terms of new images and meta-data.
- In the 3d edition Tuberculosis task was restructured to allow usage of uniform dataset, and included two subtasks - continued Severity Score (SVR) prediction subtask and a new subtask based on providing an automatic report (CT Report) on the TB case. In the 4th edition, the SVR subtask was dropped and automated CT report generation task was modified to be lung-based rather than CT-based. Because of fairly high results achieved by the participants in the CTR task in 4th edition, the task organizers have decided to discontinue the CTR task and brought back to life the Tuberculosis Type classification task from the 1st and 2nd ImageCLEFmed Tuberculosis editions to check if recent Machine Learning and Deep Learning methods allows to improve previous rather low results.
In this year edition the task is upgraded from classification problem to detection problem.
The task will include two subtasks.
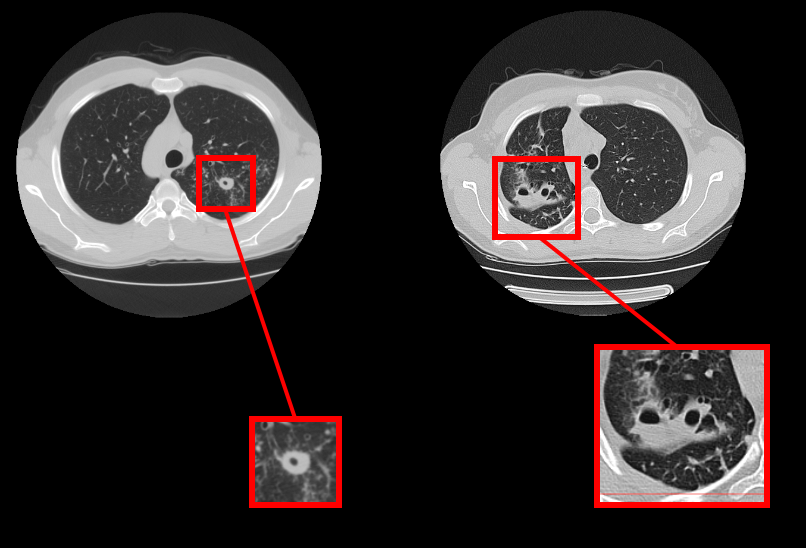
The first subtask is detection itself: participants must detect lung cavern regions in lung CT images associated with lung tuberculosis. The problem is important because even after successful treatment which fulfills the existing criteria of recovery the caverns may still contain colonies of Mycobacterium Tuberculosis that could lead to unpredictable disease relapse.
The second subtask is caverns classification problem. Participants must predict 4 binary features of caverns suggested by experienced radiologists.
The task webpages are available on AICrowd:Caverns Detection, Caverns Report(updating)
Preliminary schedule
- 15.11.2021: registration opens for all ImageCLEF tasks
- 06.02.2022: expected development data release for both subtasks
- 14.03.2022: test data release starts (depends on the task)
- 06.05.2022: deadline for submitting the participants runs (depends on the task)
- 13.05.2022: release of the processed results by the task organizers (depends on the task)
- 27.05.2022: deadline for submission of working notes papers by the participants
- 13.06.2022: notification of acceptance of the working notes papers
- 01.07.2022: camera ready working notes papers
- 5-8.09.2022: CLEF 2022, Bologna, Italy https://clef2022.clef-initiative.eu/
Task description
The goal of the first subtask (Caverns Detection) is to detect lung cavern regions in lung CT images associated with lung tuberculosis.
The of the the second subtask (Caverns Report) is to predict 4 binary features of caverns suggested by experienced radiologists.
Data
For all patients we provide a single 3D CT image with an image size per slice of 512×512 pixels and number of slices being around 100. All the CT images are stored in NIFTI file format with .nii.gz file extension (g-zipped .nii files). This file format stores raw voxel intensities in Hounsfield units (HU) as well the corresponding image metadata such as image dimensions, voxel size in physical units, slice thickness, etc. A freely-available tool called "VV" can be used for viewing the image files. Currently, there are various tools available for reading and writing NIFTI files. Among them there are load_nii and save_nii functions for Matlab and Niftilib library for C, Java, Matlab and Python, NiBabel package for Python.
Moreover, for all the CT images we provide two versions of automatically extracted masks of the lungs. These data can be downloaded together with the patients CT images. The description of the first version of segmentation can be found here. The description of the second version of segmentation can be found here. The first version of segmentation provides more accurate masks, but it tends to miss large abnormal regions of lungs in the most severe TB cases. The second segmentation on the contrary provides more rough bounds, but behaves more stable in terms of including lesion areas. In case the participants use the provided masks in their experiments, please refer to the section "Citations" at the end of this page to find the appropriate citation for the corresponding lung segmentation technique.
Evaluation methodology
(Updating)
Organizers
- Serge Kozlovski <kozlovski.serge(at)gmail.com>, Institute for Informatics, Minsk, Belarus
- Vitali Liauchuk <vitali.liauchuk(at)gmail.com>, Institute for Informatics, Minsk, Belarus
- Yashin Dicente Cid <yashin.dicente(at)warwick.ac.uk>, University of Warwick, Coventry, England, UK
- Vassili Kovalev <vassili.kovalev(at)gmail.com>, Institute for Informatics, Minsk, Belarus
- Henning Müller <henning.mueller(at)hevs.ch>, University of Applied Sciences Western Switzerland, Sierre, Switzerland
