- ImageCLEF 2024
- LifeCLEF 2024
- ImageCLEF 2023
- LifeCLEF 2023
- ImageCLEF 2022
- LifeCLEF2022
- ImageCLEF 2021
- LifeCLEF 2021
- ImageCLEF 2020
- LifeCLEF 2020
- ImageCLEF 2019
- LifeCLEF 2019
- ImageCLEF 2018
- LifeCLEF 2018
- ImageCLEF 2017
- LifeCLEF2017
- ImageCLEF 2016
- LifeCLEF 2016
- ImageCLEF 2015
- LifeCLEF 2015
- ImageCLEF 2014
- LifeCLEF 2014
- ImageCLEF 2013
- ImageCLEF 2012
- ImageCLEF 2011
- ImageCLEF 2010
- ImageCLEF 2009
- ImageCLEF 2008
- ImageCLEF 2007
- ImageCLEF 2006
- ImageCLEF 2005
- ImageCLEF 2004
- ImageCLEF 2003
- Publications
- Old resources
You are here
FungiCLEF 2023
FungiCLEF 2023

Schedule
- December 2023: registration opens
- 14 February 2023: training data release
- 24 May 2023: deadline for submission of runs by participants
- 27 May 2023: release of processed results by the task organizers
- 7 June 2023: deadline for submission of working note papers by participants [CEUR-WS proceedings]
- 30 June 2023: notification of acceptance of working note papers [CEUR-WS proceedings]
- 7 July 2023: camera ready copy of participant's working note papers and extended lab overviews by organizers
- 18-21 Sept 2023: CLEF 2023 Thessaloniki
Results
All teams that provided a runnable code were scored, and their scores are available on HuggingFace.
Official competition results and intermediate validation results are listed in the Private and Public leaderboards, respectively.
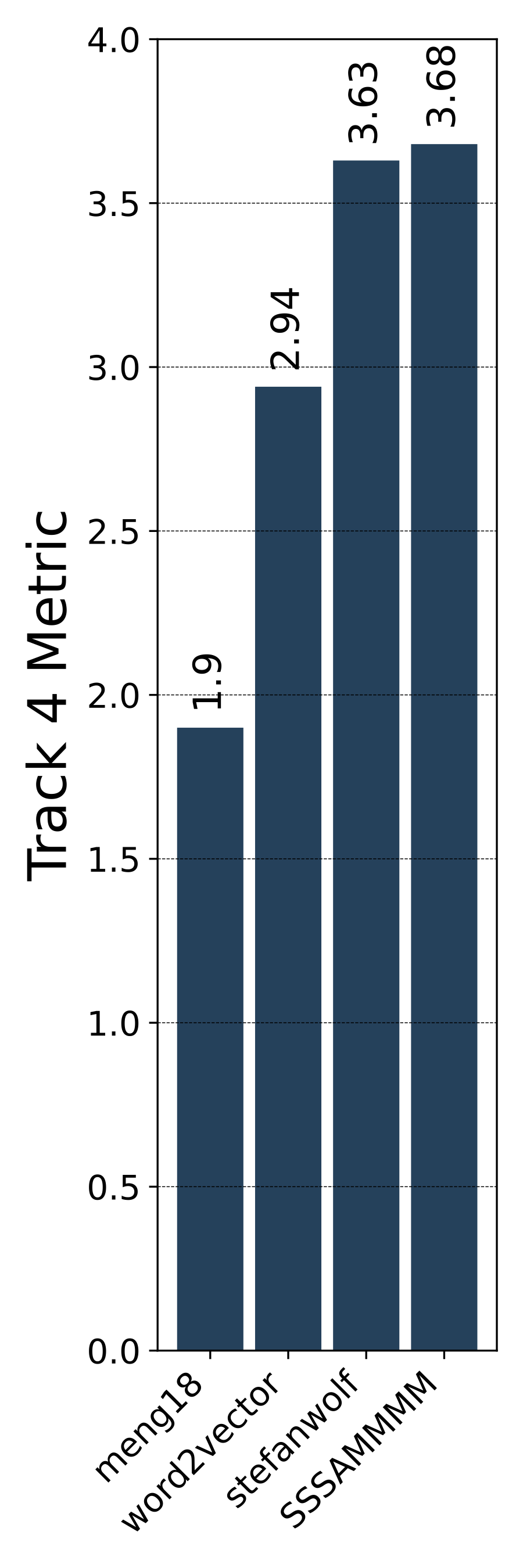
- The 1st place team: meng18 (Huan Ren, Han Jiang, Wang Luo, Meng Meng and Tianzhu Zhang)
- The 2nd place team: stefanwolf (Stefan Wolf and Jürgen Beyerer)
- The 3rd place team: word2vector (Feiran Hu, Peng Wang, Yangyang Li, Chenlong Duan, Zijian Zhu, Yong Li and Xiu-Shen Wei)
Public Leaderboard Evaluation
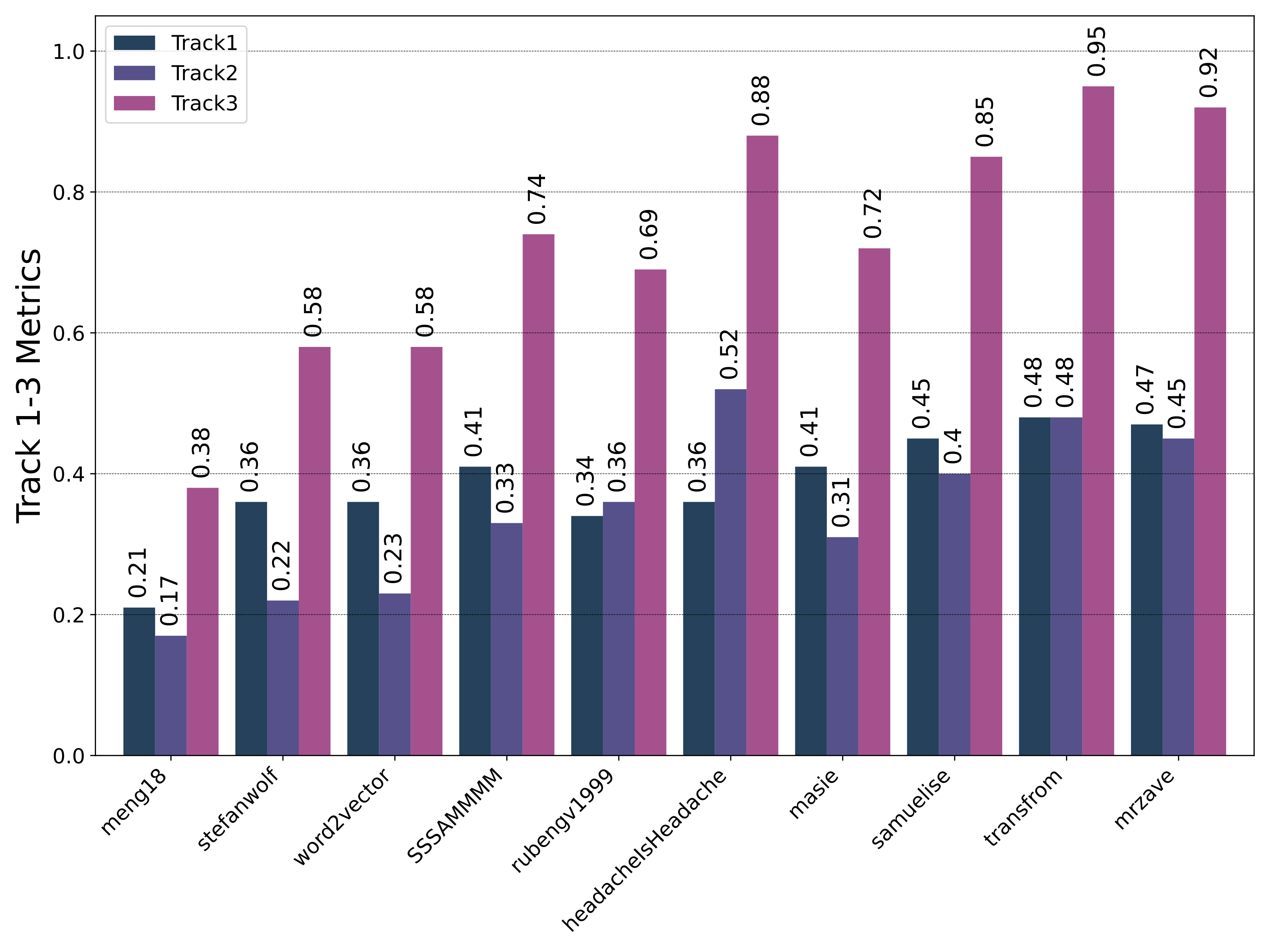
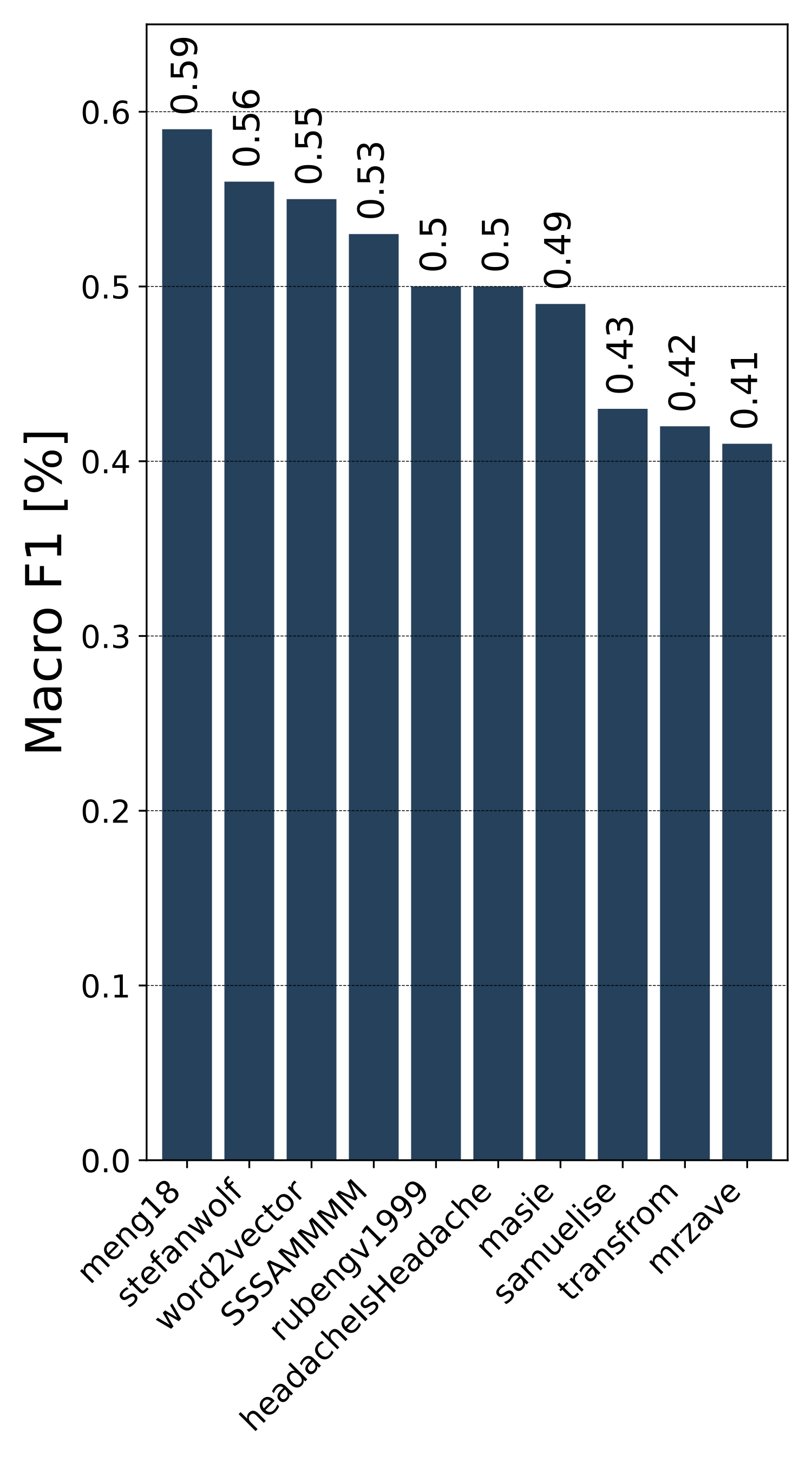
Private Leaderboard Evaluation
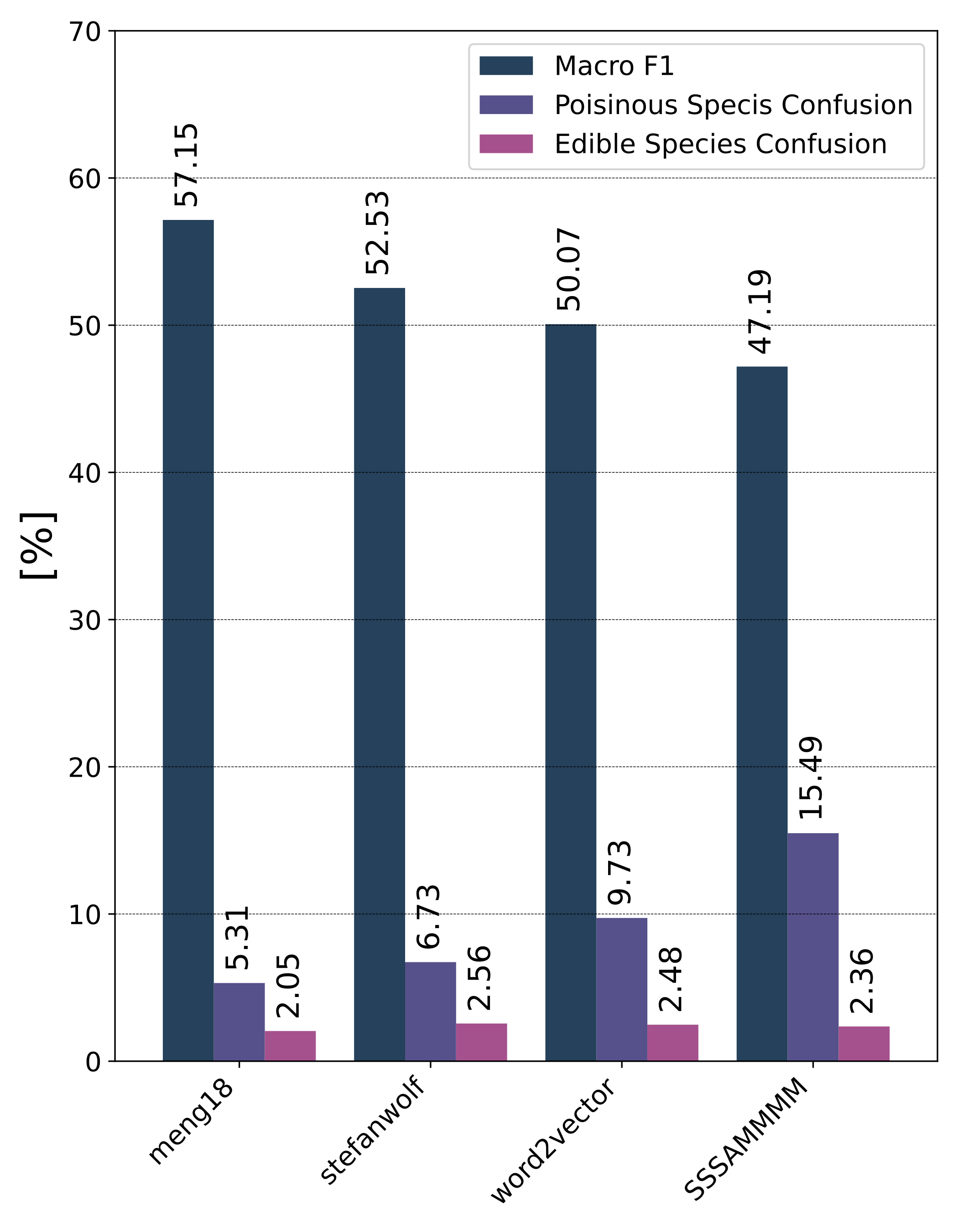
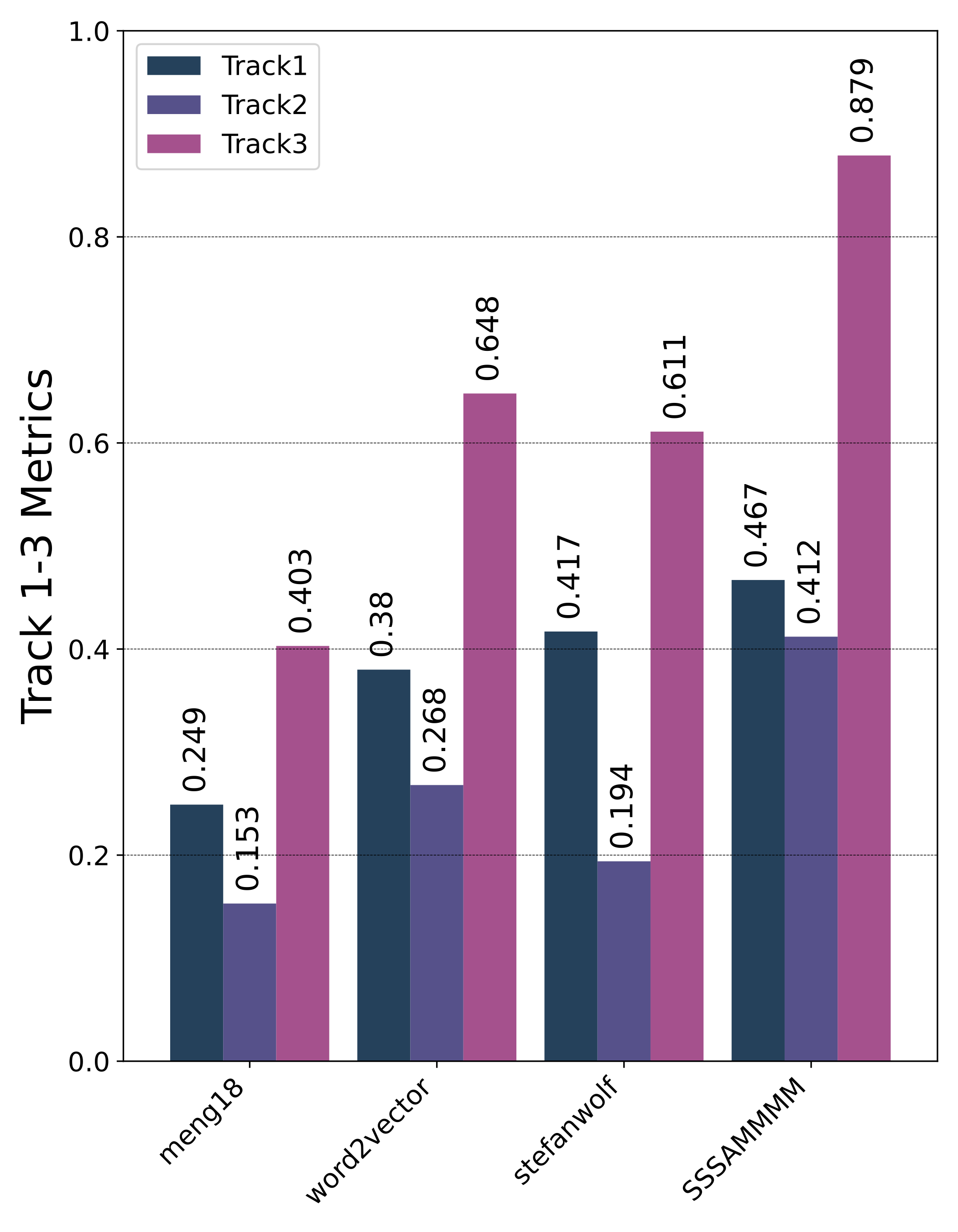
Motivation
Automatic recognition of fungi species assists mycologists, citizen scientists and nature enthusiasts in species identification in the wild. Its availability supports the collection of valuable biodiversity data. In practice, species identification typically does not depend solely on the visual observation of the specimen but also on other information available to the observer - such as habitat, substrate, location and time. Thanks to rich metadata, precise annotations, and baselines available to all competitors, the challenge provides a benchmark for image recognition with the use of additional information. Moreover, the toxicity of a mushroom can be crucial for the decision of a mushroom picker. We will explore the decision process within the competition beyond the commonly assumed 0/1 cost function.
Task Description
Given the set of real fungi species observations and corresponding metadata, the goal of the task is to create a classification model that, for each observation (multiple photographs of the same individual + geographical location), returns a ranked list of predicted species. The classification model will have to fit limits for memory footprint (ONNX model with max size of 1GB) and prediction time limit (will be announced later) measured on the submission server. The model should have to consider and minimize the danger to human life, i.e., the confusion between poisonous and edible species.
Participation requirements
- 1. Subscribe to CLEF (LifeCLEF – SnakeCLEF2023 task) by filling out this form.
- 2. Submit your solution before the competition deadline. More info on a HuggingFace competition platform.
Data
The challenge dataset is primarily based on the data from the Danish Fungi 2020 dataset, which contains 295,938 training images belonging to 1,604 species observed mostly in Denmark.
All training samples passed an expert validation process, guaranteeing high-quality labels. Rich observation metadata about habitat, substrate, time, location, EXIF etc. are provided.
The validation set contains 30,131 observations with 60,832 images and 2,713 species, covering the whole year and including observations collected across all substrate and habitat types. The public test set contains 30,130 observations with 60,225 images and similar data distribution as the validation set.
Using additional data or metadata is permitted!
| Image Data | Metadata |
Other resources
- Papers describing the Danish Fungi dataset and our Fungi Recognition App.
- CVPR - FGVC workshop
- LifeCLEF
page for more information about the FGVC10 workshop.
page for more information about LifeCLEF challenges and working notes submission procedure.
Publication Track
LifeCLEF 2023 is an evaluation campaign that is being organized as part of the CLEF initiative labs. The campaign offers several research tasks that welcome team participation worldwide.
The results of the campaign appear in the working notes proceedings published by CEUR Workshop Proceedings (CEUR-WS.org).
Selected contributions among the participants will be invited for publication in the following year in the Springer Lecture Notes in Computer Science (LNCS), together with the annual lab overviews.
Context
This competition is held jointly as part of:
- the LifeCLEF 2023 lab of the CLEF 2023 conference, and of
- the FGVC10 workshop, organized in conjunction with CVPR 2023
The participants are required to participate in the LifeCLEF lab by registering for it using the CLEF 2023 labs registration form (and checking "Task 5 - SnakeCLEF" of LifeCLEF).
Only registered participants should submit a working-note paper to peer-reviewed LifeCLEF proceedings (CEUR-WS) after the competition ends."
This paper should provide sufficient information to reproduce the final submitted runs.
Only participants who submitted a working-note paper will be part of the officially published ranking used for scientific communication.
Organizers
- Lukas Picek, PiVa AI & Dept. of Cybernetics, FAV, University of West Bohemia, Czechia, lukaspicek@gmail.com
- Milan Sulc, Czechia, milansulc01@gmail.com
- Rail Chamidullin, PiVa AI & Dept. of Cybernetics, FAV, University of West Bohemia, Czechia
- Jiri Matas, The Center for Machine Perception Dept. of Cybernetics, FEE, Czech Technical University, Czechia, matas@cmp.felk.cvut.cz
- Jacob Heilmann-Clausen, Center for Macroecology, Evolution and Climate; University of Copenhagen, Denmark, jheilmann-clausen@sund.ku.dk
Machine Learning
Mycology
Credits
Acknowledgement





