- ImageCLEF 2024
- LifeCLEF 2024
- ImageCLEF 2023
- LifeCLEF 2023
- ImageCLEF 2022
- LifeCLEF2022
- ImageCLEF 2021
- LifeCLEF 2021
- ImageCLEF 2020
- LifeCLEF 2020
- ImageCLEF 2019
- LifeCLEF 2019
- ImageCLEF 2018
- LifeCLEF 2018
- ImageCLEF 2017
- LifeCLEF2017
- ImageCLEF 2016
- LifeCLEF 2016
- ImageCLEF 2015
- LifeCLEF 2015
- ImageCLEF 2014
- LifeCLEF 2014
- ImageCLEF 2013
- ImageCLEF 2012
- ImageCLEF 2011
- ImageCLEF 2010
- ImageCLEF 2009
- ImageCLEF 2008
- ImageCLEF 2007
- ImageCLEF 2006
- ImageCLEF 2005
- ImageCLEF 2004
- ImageCLEF 2003
- Publications
- Old resources
You are here
SnakeCLEF 2023
SnakeCLEF 2023

Tentative Schedule
- December 2023: Registration opens
- 14 February 2023: training data release
- 24 May 2023: deadline for submission of runs by participants
- 27 May 2023: release of processed results by the task organizers
- 7 June 2023: deadline for submission of working note papers by participants [CEUR-WS proceedings]
- 30 June 2023: notification of acceptance of working note papers [CEUR-WS proceedings]
- 7 July 2023: camera-ready copy of participant's working note papers and extended lab overviews by organizers
- 18-21 Sept 2023: CLEF 2023 Thessaloniki
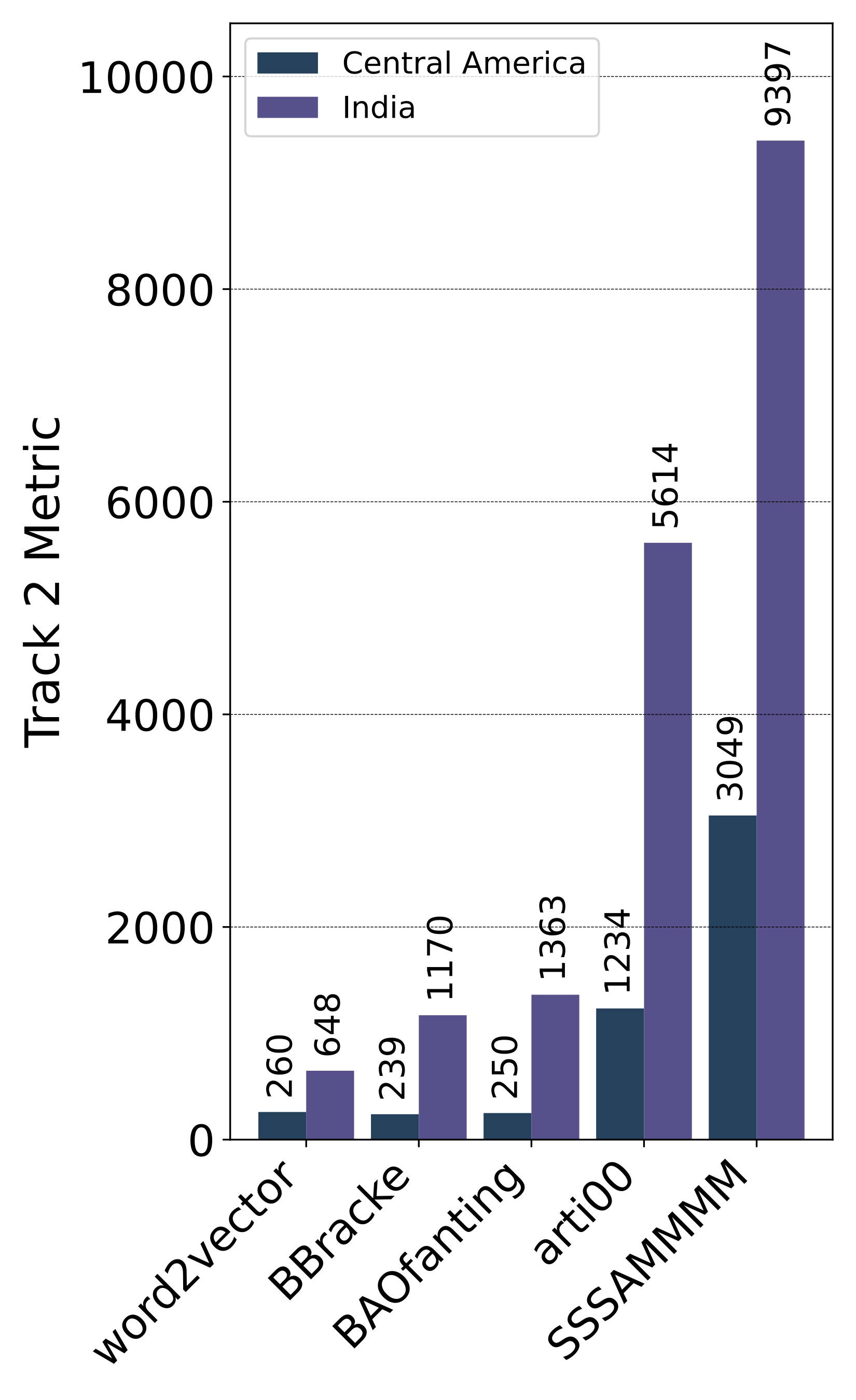
Results
All teams that provided a runnable code were scored, and their scores are available on HuggingFace.
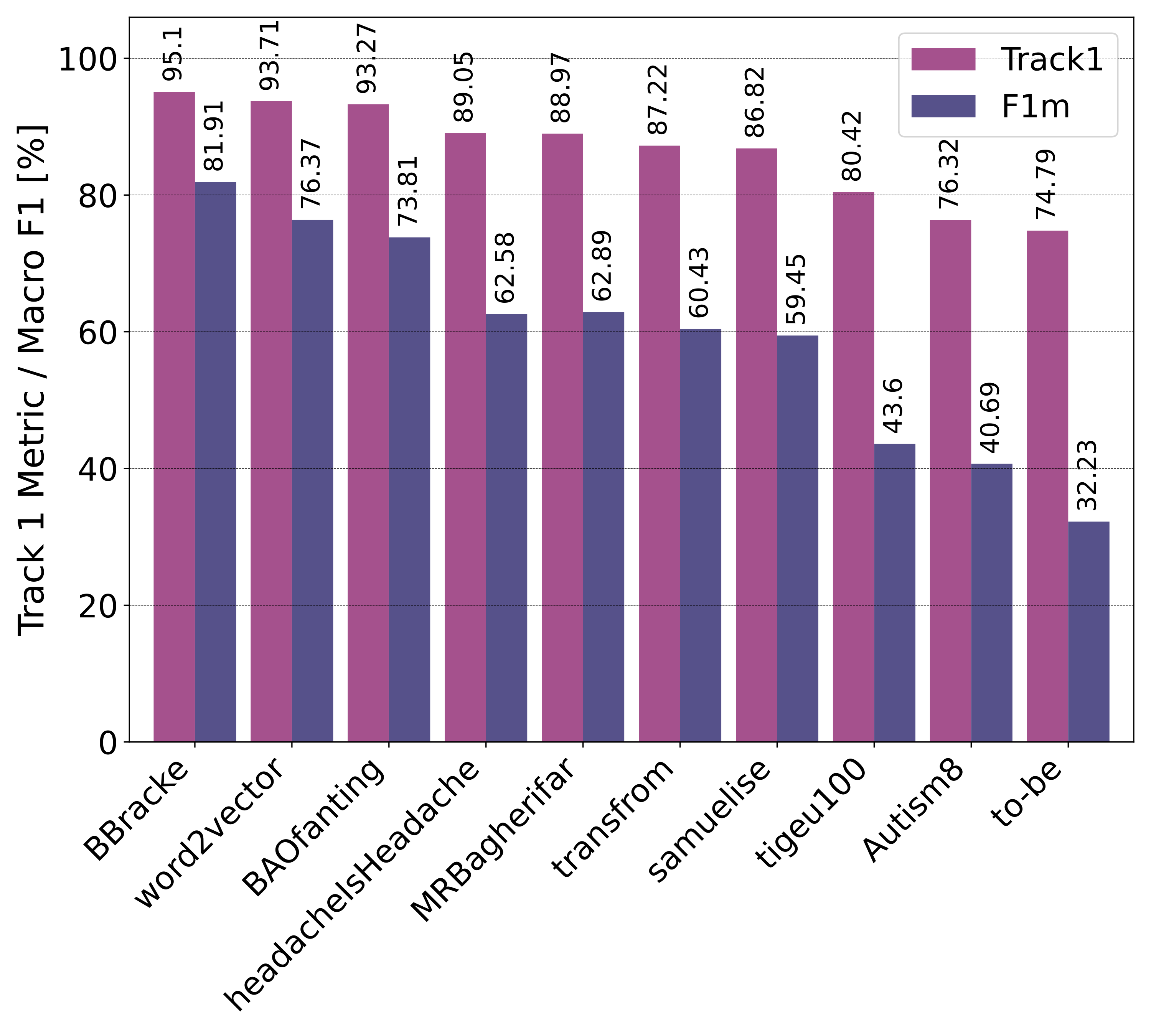
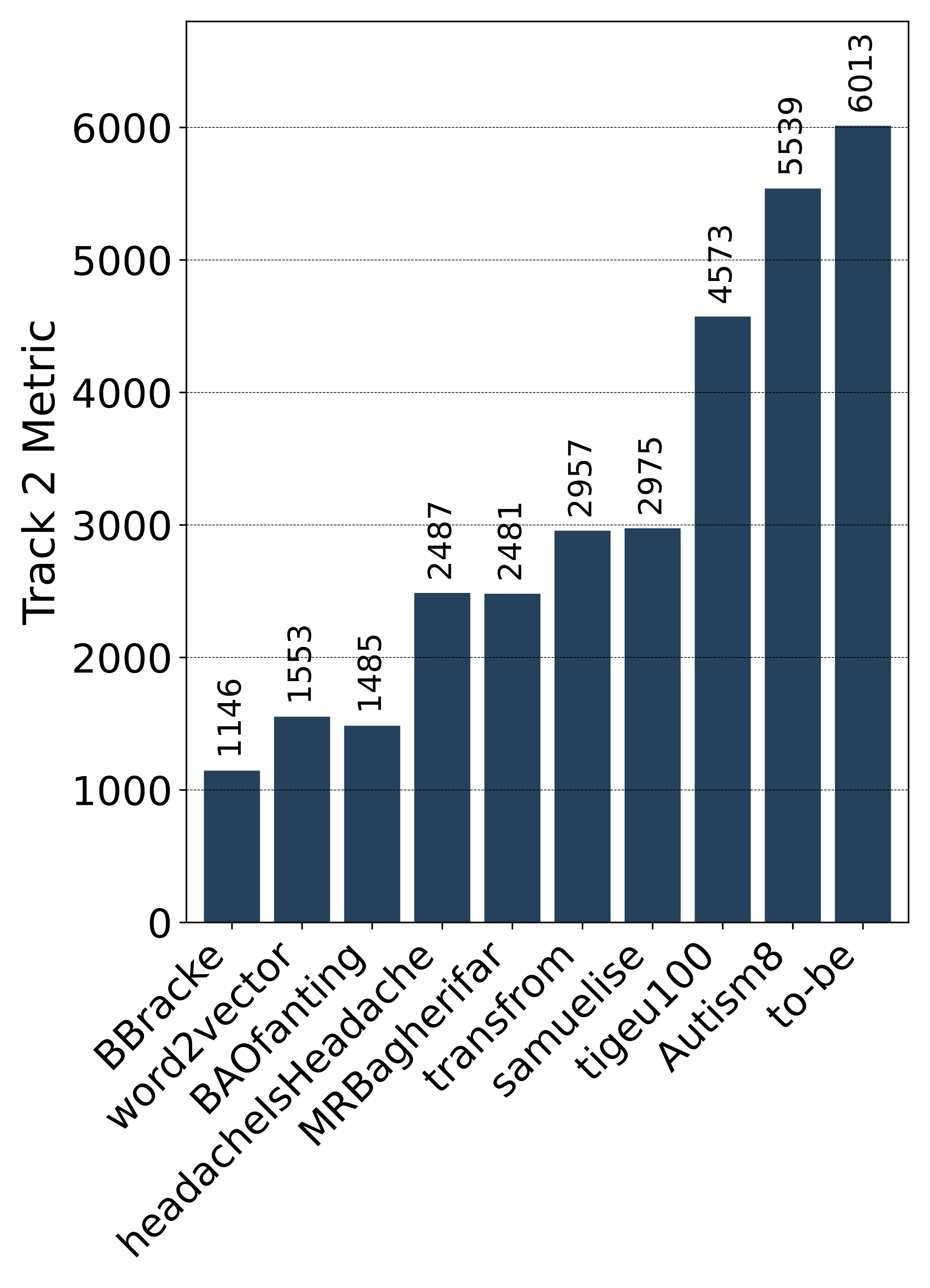
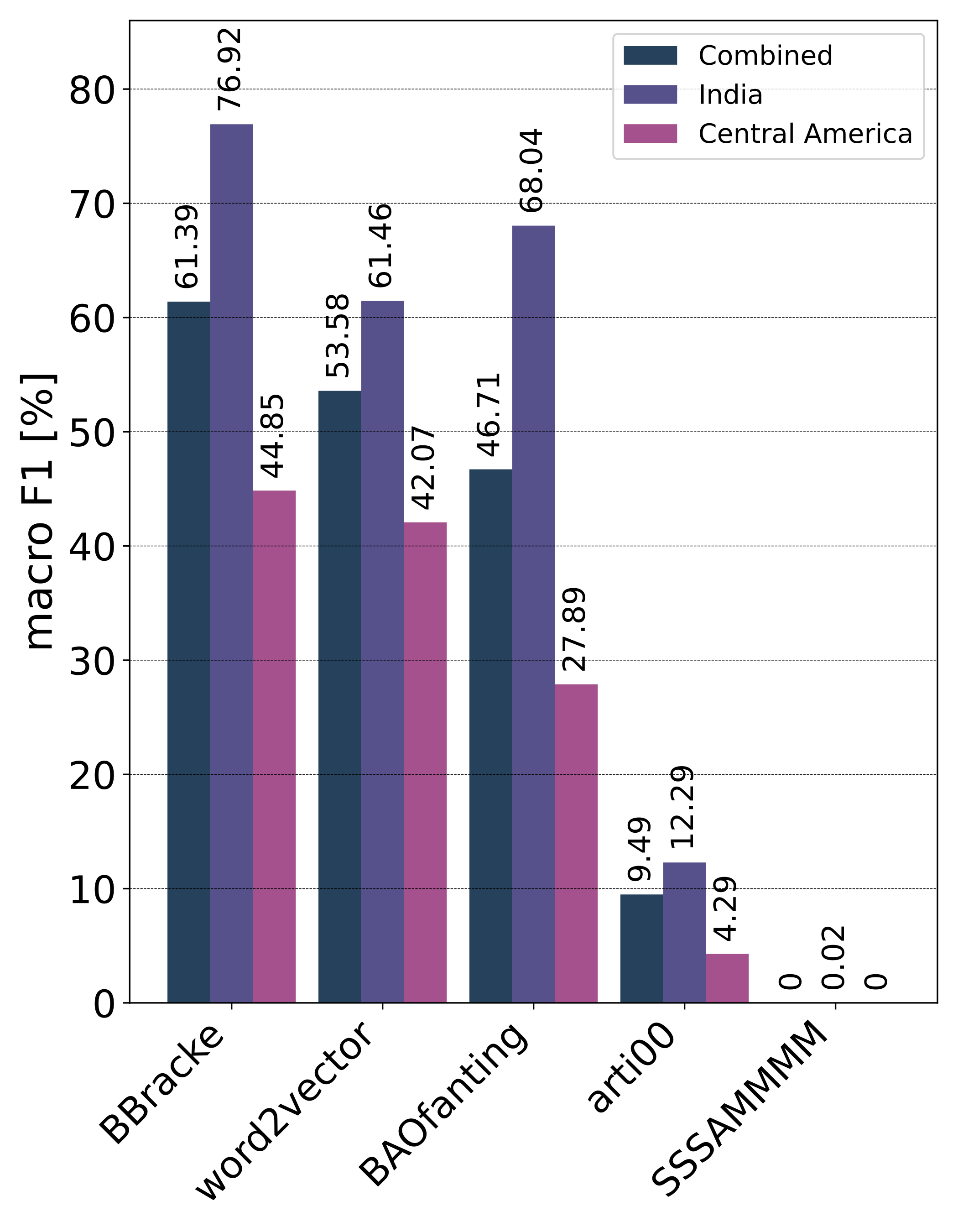
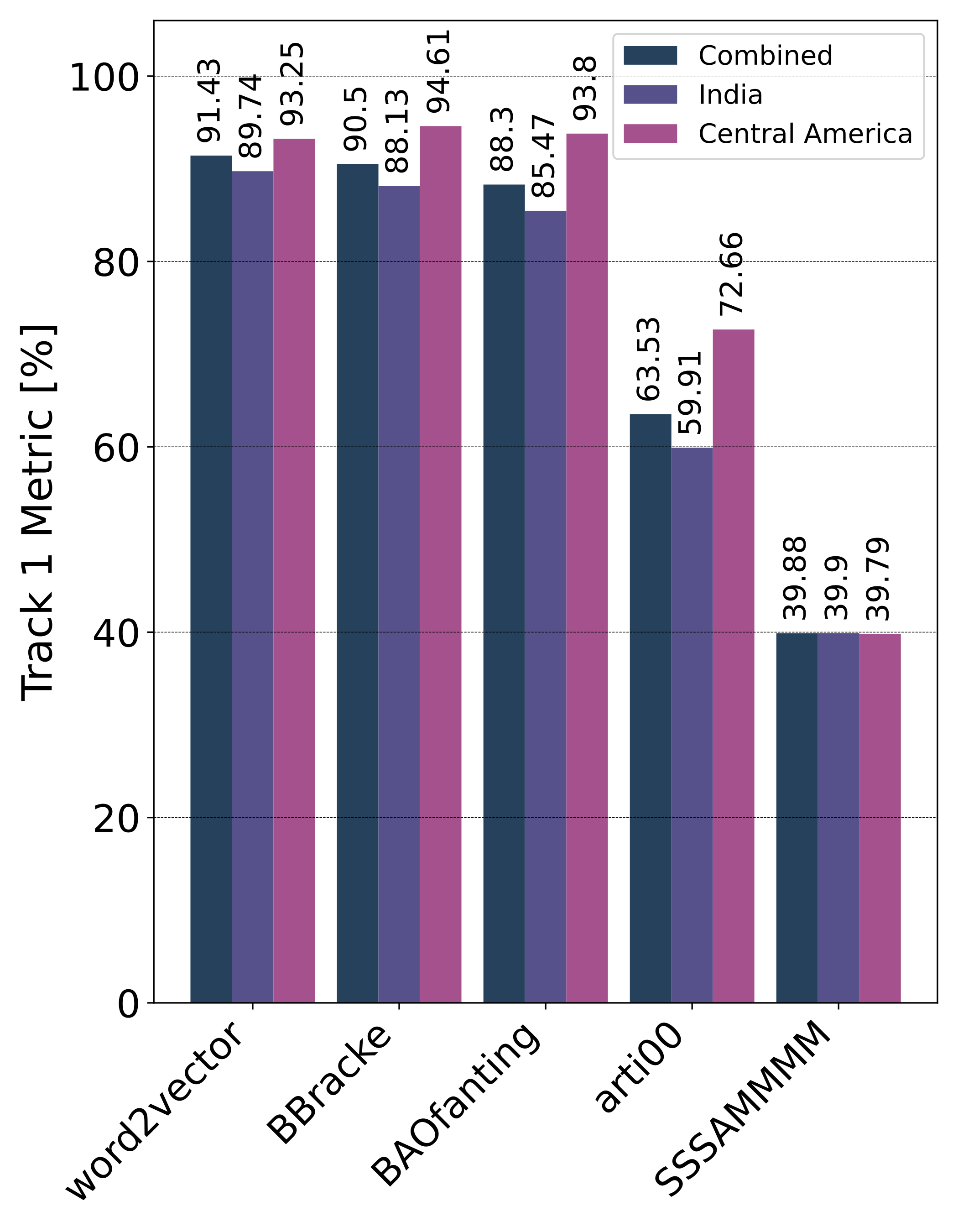
Official competition results and intermediate validation results are listed in the Private and Public leaderboards, respectively.
- The 1st place team: word2vector (Feiran Hu, Peng Wang, Yangyang Li, Chenlong Duan, Zijian Zhu, Fei Wang, Faen Zhang, Yong Li and Xiu-Shen Wei)
- The 2nd place team: BBracke (Benjamin Bracke, Mohammadreza Bagherifar, Louise Bloch and Christoph M. Friedrich)
- The 3rd place team: BAOfanting (Zhennan Shi, Huazhen Chen, Chang Liu and Jun Qiu)
Public Leaderboard Evaluation
Private Leaderboard Evaluation
Motivation
Developing a robust system for identifying species of snakes from photographs is an important goal in biodiversity but also for human health. With over half a million victims of death & disability from venomous snakebite annually, understanding the global distribution of the > 4,000 species of snakes and differentiating species from images (particularly images of low quality) will significantly improve epidemiology data and treatment outcomes. We have learned from previous editions that “machines” can accurately recognize (F1 = 86,5% and Top1 Accuracy = 94%) even in scenarios with long-tailed distributions and around 1,600 species. Thus, testing over real Medically Important Scenarios and specific countries (primarily tropical and subtropical) and integrating the medical importance of species is the next step that should provide a more reliable machine prediction.
The difficulty of snake species identification – from both a human and a machine perspective – lies in the high intra-class and low inter-class variance in appearance, which may depend on geographic location, colour morph, sex, or age. At the same time, many species are visually similar to other species (e.g. mimicry).
Our knowledge of which snake species occur in which countries is incomplete, and it is common that most or all images of a given snake species might originate from a small handful of countries or even a single country. Furthermore, many snake species resemble species found on other continents, with which they are entirely allopatric. Knowing the geographic origin of an unidentified snake can narrow down the possible correct identifications considerably. In no location on Earth do more than 126 of the approximately 4,000 snake species co-occur. Thus, regularization to all countries is a critical component of any snake identification method.
Task Description
Given the set of authentic snake species observations – multiple photographs of the same individual – and corresponding geographical locations, the goal of the task is to create a classification model that returns a ranked list of species. The classification model will have to fit limits for memory footprint (ONNX model with max size of 1GB) and prediction time limit (will be announced later) measured on the submission server. The model should have to consider and minimize the danger to human life and the waste of antivenom if a bite from the snake in the image were treated as coming from the top-ranked prediction.
Participation requirements
- 1. Subscribe to CLEF (LifeCLEF – SnakeCLEF2023 task) by filling out this form.
- 2. Submit your solution before the competition deadlinel. More info on a HuggingFace competition platform.
Data
For this year's challenge, we prepared a development dataset based on 103,404 snake observations with 182,261 photographs belonging to 1,784 snake species and observed in 214 countries. The data were gathered from the online biodiversity platforms -- iNaturalist and Herpmapper. The provided dataset has a heavy long-tailed class distribution, where the most frequent species (Natrix natrix) is represented by 1,262 observations (2,079 images) and the least frequent species by just 3 observations.
Using additional data or metadata is permitted!
| Image Data (Training set) | Image Data (Validation and Test sets) | Metadata |
Evaluation process
This competition provides an evaluation ground for the development of methods suitable for not just snake species recognition. We want you to evaluate new bright ideas rather than finishing first on the leaderboard. Thus, we this year we will award an authorship / co-authorship on a Journal publication and payment for an OpenAccess fee.
The whole evaluation process will be divided into two parts (i) CSV-based evaluation on EvalAI, and (ii) ONNX model evaluation on our private data.
The final performance will be provided only on the LifeCLEF web.
The test set (private data) will consider different medically important scenarios and overlooked regions.
Evaluation Platforms: EvalAI, Kaggle (Links will be announced.)
Test Platform: TBA
The final evaluation will be conducted on provided ONNX models. The ONNX model will have to have <1 GB and will have to be uploaded together with a script for running the inference (run.py). The final evaluation will use secret test data. We want to encourage the teams to upload compact models with fast runtimes, aiming at real-world applications. We discourage the usage of complex ensembles that do not have a significant impact on the results. This way, we hope that participants will focus on novel approaches rather than combining all known architectures. Also, this setup is suitable for the experimental reproduction of your final systems and as such will help the whole community the most.
Metrics: This year we will calculate various metrics. First, we will calculate standard Acc and macro averaged F1. Besides, we will calculate venomous species confusion error, i.e., a number of samples with venomous species confused for harmless and divided by the number of venomous species in the test set.
To motivate research in recognition scenarios with uneven costs for different errors, such as mistaking a venomous snake for a harmless one, this year's challenge goes beyond the 0-1 loss common in classification. We make some assumptions to reduce the complexity of the evaluation. We consider that there exists a universal antivenom that is applicable to all venomous snake bites. Furthermore, such antivenom is not harmful when applied to a healthy human. Hence, we will penalize the misclassification of a venomous species with a harmless one but not the other way around. Although this solution is not perfect, it is a first step into a more complex evaluation of snake bites.
Let us consider a function \(p\) such that \(p(s)=1\) if species \(s\) is venomous, otherwise \(p(s)=0\).
For a correct species \(y\) and predicted species \(\hat y\), the loss \(L(y, \hat y)\) is given as follows:
\[L(y, \hat y) = \left\{
\begin{array}{ll}
0 & \text{ if } y = \hat y \\
1 & \text{ if } y \neq \hat y \text{ and } p(y)=0 \text{ and } p(y)=0 \\
2 & \text{ if } y \neq \hat y \text{ and } p(y)=0 \text{ and } p(\hat y)=1 \\
2 & \text{ if } y \neq \hat y \text{ and } p(y)=1 \text{ and } p(\hat y)=1 \\
5 & \text{ if } y \neq \hat y \text{ and } p(y)=1 \text{ and } p(\hat y)=0 \\
\end{array}
\right.
\]
Note: The costs were selected to illustrate a higher cost when a venomous snake is mistaken for a harmless one. We do not claim the selected costs reflect the risks in a practical scenario: practical costs would have to be determined by assessing what exactly follows after the recognition process. One can imagine several aspects, e.g., the health risk of the venom, the cost of the antivenom and so on.
The challenge metric is the sum of \(L) over all test observations:
\[ \mathbf L = \sum_i L(y_i, \hat y_i)
\]
The other one will include the overall classification rate (F1) and the venomous species confusion error. The
metric is a weighted average between the macro F1-score and the weighted accuracies of different types of confusion.
\[
M = (w_1 F_1 + w_2 (100-P_1) + w_3 (100-P_2) + w_4 (100-P_3) + w_5 (100-P_4)) / \sum_i^5 w_i,
\]
where \(w_1=1.0, w_2=1.0, w_3=2.0. w_4=5.0. w_5=2.0\) are the weights of individual terms. \(F_1\) is the macro F1-score,
\(P_1\) is the percentage of wrongly classified harmless species as another harmless species,
\(P_2\) is the percentage of wrongly classified harmless species as another venomous species,
\(P_3\) is the percentage of wrongly classified venomous species as another harmless species, and
\(P_4\) is the percentage of wrongly classified venomous species as another venomous species.
This metric has a lower bound of 0% and an upper bound of 100%. Lower bound is achieved if you misclassify
every species, and furthermore, you misclassify every harmless species as a venomous one and vice-versa.
On the other hand, if F1 is 100% (every species is classified correctly), every \(P_i\) must be equal to zero by
definition, and 100% will be achieved.
Other Resources
For more SnakeCLEF-related info, please refer to overview papers from previous editions. Besides, you can check out other competitions from the CLEF-LifeCLEF and CVPR-FGVC workshops.
- SnakeCLEF2022 Overview Paper, SnakeCLEF2021 Overview Paper, and SnakeCLEF2020 Overview Paper
- CVPR - FGVC workshop page for more information about the FGVC10 workshop.
- LifeCLEF page for more information about LifeCLEF challenges and working notes submission procedure.
Publication Track
LifeCLEF 2023 is an evaluation campaign that is being organized as part of the CLEF initiative labs. The campaign offers several research tasks that welcome team participation worldwide.
The results of the campaign appear in the working notes proceedings published by CEUR Workshop Proceedings (CEUR-WS.org).
Selected contributions among the participants will be invited for publication in the following year in the Springer Lecture Notes in Computer Science (LNCS), together with the annual lab overviews.
Context
This competition is held jointly as part of:
- the LifeCLEF 2023 lab of the CLEF 2023 conference, and of
- the FGVC10 workshop, organized in conjunction with CVPR 2023
The participants are required to participate in the LifeCLEF lab by registering for it using the CLEF 2023 labs registration form (and checking "Task 5 - SnakeCLEF" of LifeCLEF).
Only registered participants should submit a working-note paper to peer-reviewed LifeCLEF proceedings (CEUR-WS) after the competition ends."
This paper should provide sufficient information to reproduce the final submitted runs.
Only participants who submitted a working-note paper will be part of the officially published ranking used for scientific communication.
Organizers
- Lukas Picek, PiVa AI & Dept. of Cybernetics, FAV, University of West Bohemia, Czechia, lukaspicek@gmail.com
- Marek Hruz, Dept. of Cybernetics, FAV, University of West Bohemia, Czechia, mhruz@ntis.zcu.cz
- Rail Chamidullin, PiVa AI & Dept. of Cybernetics, FAV, University of West Bohemia, Czechia
- Andrew Durso, Department of Biological Sciences, Florida Gulf Coast University, Fort Myers, USA, amdurso@gmail.com
- Isabelle Bolon, Institute of Global Health, Department of Community Health and Medicine, University of Geneva, Switzerland
Machine Learning
Herpatology
Clinical Expert
Credits
Acknowledgement






